Background
Traditionally small molecules have been used to inhibit the action of a target protein by occupying and blocking a functional region of the protein. An alternative innovative approach is the development of proteolysis targeting chimeras (PROTACs), i.e. heterobifunctional compounds consisting of one moiety that binds a Cullin RING E3 ubiquitin ligase linked to another that binds a desired protein of interest (POI), bringing the ligase and the POI into close spatial proximity (Fig. 1). This hijacks the intrinsic catalytic activity of the E3 ligase from the natural and directs it toward the POI as a neo-substrate, triggering its poly-ubiquitination and subsequent proteasome-dependent degradation (for a review of E3 ligase degron recognition, see Lucas & Ciulli, Curr. Opin Struct. Biol. 2017).
As a result, a PROTAC acts as a degrader of the target as opposed to just an inhibitor, enabling the effective post-translational elimination of a target gene product in living organisms. This approach presents many advantages compared to conventional target inhibition. One of the most attractive features of the approach is that a PROTAC molecule acts sub-stoichiometrically, i.e. it only needs to bind a molecule of target once to induce its degradation, and then is released and set free to bind another molecule of target and carry on, as in a catalytic cycle [1]. For this reason, the concentrations required for PROTACs to be active in cells tend to be much lower compared to those needed to be reached and maintained with inhibitors, which can lead to fewer off-target effects and a more selective chemical intervention on the desired target.
Research
After a decade of minor advances since its first conceptualization [2], recent discoveries from us and others [3,4] of low-molecular weight, high-affinity and selective ligands for E3 Cullin RING ligases with crystallographically defined binding modes have fully enabled the universal applicability of this approach [5]. In a recently published article [6], the Ciulli lab has shown that the Bromo- and Extra-terminal (BET) pan-selective inhibitor JQ1 could be transformed into a PROTAC compound that unexpectedly led to effective and preferential removal of a single member of the protein family, Brd4, leaving the homologous BET members Brd2 and Brd3 untouched. This was achieved by conjugating JQ1 to a potent ligand for the VHL E3 ligase that we had previously discovered [3], via a PEG linker, resulting in PROTAC compound MZ1 (Fig. 2). MZ1 is active at a sufficiently low concentration not to induce stabilization and transcriptional activation of HIF-α, the natural substrate of VHL. Brd4 selectivity was consistently observed in all the cell lines and with all the compounds tested [6]. In contrast, tethering the same pan-BET inhibitor to a ligand for a different E3 ligase, cereblon, led to pan-selective degradation of all BET proteins [7,8]. BET targeting PROTACs show pronounced and long-lasting anti-proliferative effect against leukemia cells.
Our discovery of MZ1 enables Brd4-specific responses in cancer cells to be properly assessed, which will have a major impact in the field of epigenetic gene regulation and cancer. More generally, it shows for the first time that non-selective or promiscuous inhibitors can be turned in to exquisitely selective degraders, with negligible off-target effects and improved therapeutic potential. Building on from these observations, we have recently solved a breakthrough first crystal structure of MZ1 bound to both its target Brd4 and E3 ligase VHL [9]. Our structure shows how MZ1 folds into itself, allowing Brd4 and VHL to make extensive contact with each other. Further computational and biophysical studies in solution contributed to establish the mechanistic and structural basis for PROTAC selectivity, shining unprecedented new light into how PROTACs work. Guided by the crystal structure, new degrader AT1 showing improved selectivity for Brd4 degradation was discovered [9].
The lab is interested in assessing broad applicabilities of selective target degradation being applied to other target classes. We are also actively engaged with pharmaceutical partners to explore the exciting therapeutic opportunities provided by PROTACs.
- Bondeson et al. Nat Chem Biol 11, 611-617 (2015).
- Sakamoto, K. M. et al. P Natl Acad Sci Usa 98, 8554–8559 (2001).
- Galdeano, C. et al. J Med Chem 57, 8657–8663 (2014); Van Molle I. et al., Chem Biol 19, 1300–12 (2012); Buckley D. L. et al. Angew Chem Intl Ed 51, 11463-11467 (2012); Buckley D. L. et al., J Am Chem Soc 134, 4465–68 (2012).
- Fischer, E. S. et al. Nature 512, 49–53 (2014).; Chamberlain, P. P. et al. Nat Struct Mol Biol 21, 803–809 (2014).
- Deshaies, R. J. Nat Chem Biol 11, 634–635 (2015).
- Zengerle, M., Chan, K.-H. & Ciulli, A. ACS Chem Biol 10, 1770–1777 (2015).
- Winter, G. E. et al. Science 348, 1376–1381 (2015).
- Lu, J. et al. Chem Biol 22, 755–763 (2015).
- Gadd, M.S. et al. Nat Chem Biol (2017), Article ASAP
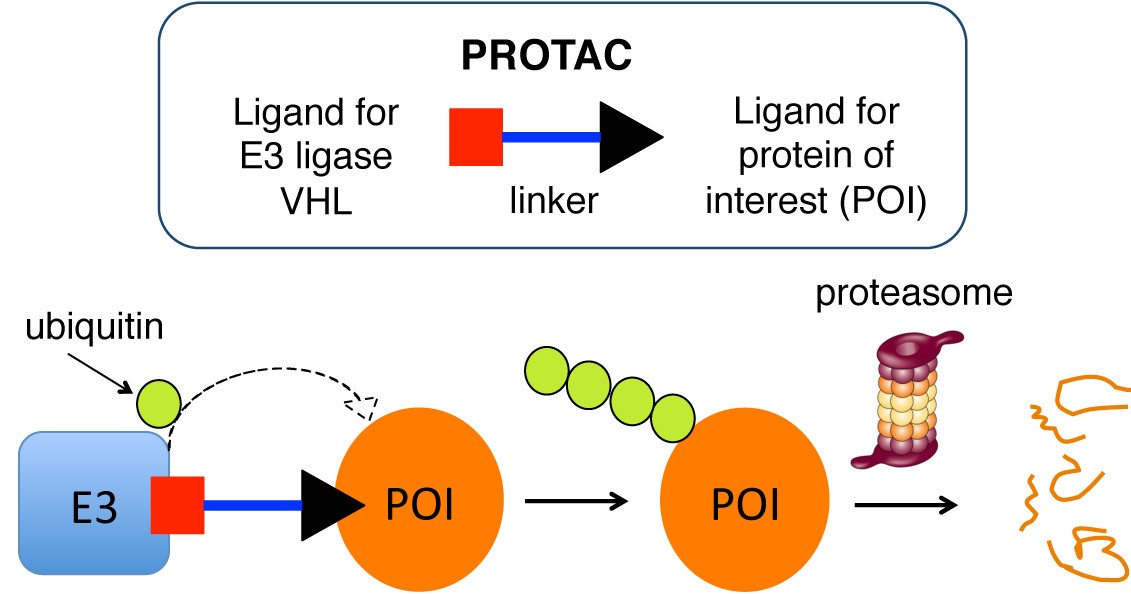
Figure 1. Schematic of the PROTAC approach. A PROTAC is a heterobifunctional molecule comprising of an E3 ligase ligand (red square) joined to a ligand for a protein of interest (POI, black triangle) via a linker unit (blue bar). PROTACs bring the E3 ligase (light blue) and the POI (orange circle) in close proximity with each other, triggering multiple transfer of ubiquitin (green circle) to the target, which ultimately leads to the proteasomal degradation of the POI inside a cell.
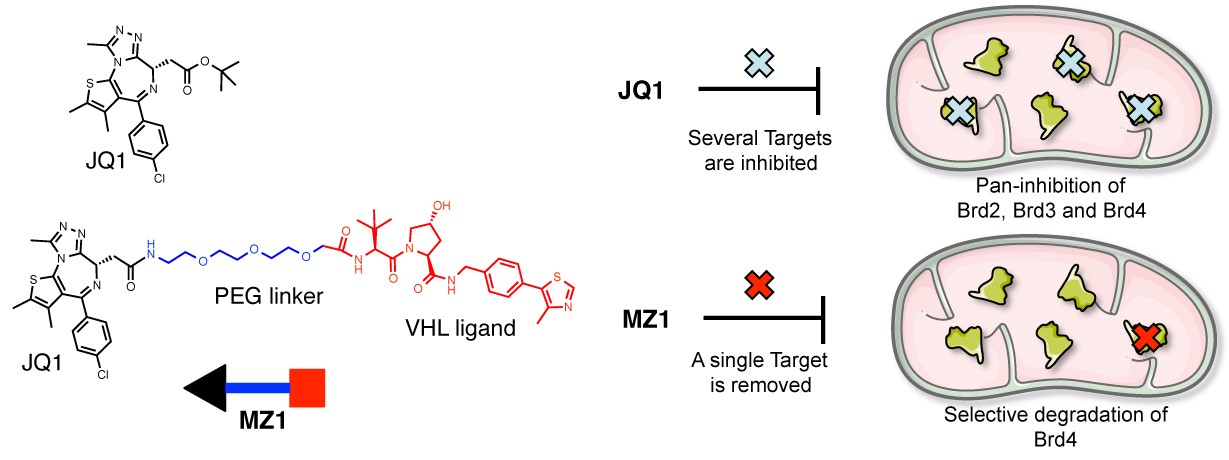
Figure 2. PROTAC compound MZ1 is a selective Brd4 degrader. MZ1 is obtained by conjugating BET inhibitor JQ1 (black) to a potent VHL ligand (red) using a short PEG linker (blue). Although JQ1 is a pan-selective inhibitor of all BET proteins Brd2, Brd3 and Brd4, MZ1 preferentially induces degradation of Brd4 alone.
Selected references: Research Articles
14. Imaide, Satomi; Riching, Kristin M.; Vetma, Vesna; Whitworth, Claire; Hughes, Scott J.; Trainor, Nicole; Mahan, Sarah D.; Murphy, Nancy; Chan, Kwok-Ho; Testa, Andrea; Maniaci, Chiara; Urh, Marjeta; Daniels, Danette L.*; Ciulli, Alessio*
Trivalent PROTACs Enhance Protein Degradation Through Cooperativity and Avidity
ChemRxiv. Preprint posted on 12 Nov 2020. DOI: 10.26434/chemrxiv.13218695.v1
13. Klein, V.G., Townsend, C.E., Testa, A., Zengerle, M., Maniaci, C., Hughes, S.J., Chan, K.-H., Ciulli, A., Lokey, R.S.
Understanding and improving the membrane permeability of VH032-based PROTACs
ACS Med. Chem. Lett. 2020, 11 (9), 1732-1738
12. Testa, A., Hughes, S.J., Lucas, X., Wright, J.E., Ciulli, A.*
Structure-Based Design of a Macrocyclic PROTAC
Angew. Chem. Int. Ed. 2020, 59 (4), 1727-1734
11. Tovell, H., Testa, A., Zhou, H., Shpiro, N., Crafter, C., Ciulli, A.*, Alessi, D.R.*
Design and characterization of SGK3-PROTAC1, an isoform specific SGK3 kinase PROTAC degrader
ACS Chem. Biol. 2019, 14 (9), 2024-2034
10. Farnaby W, Koegl M, Roy MJ, Whitworth C, Diers E, Trainor N, Zollman D, Steurer S, Karolyi-Oezguer J, Riedmueller C, Gmaschitz T, Wachter J, Dank C, Galant M, Sharps B, Rumpel K, Traxler E, Gerstberger T, Schnitzer R, Petermann O, Greb P, Weinstabl H, Bader G, Zoephel A, Weiss-Puxbaum A, Ehrenhöfer-Wölfer K, Wöhrle S, Boehmelt G, Rinnenthal J, Arnhof H, Wiechens N, Wu MY, Owen-Hughes T, Ettmayer P, Pearson M, McConnell DB*, Ciulli A.*
BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design
Nat. Chem. Biol. 2019, 15 (7), 672-680
9. Tovell, H., Testa, A., Maniaci, C., Zhou, H., Prescott, A.R., Macartney, T., Ciulli, A.*, Alessi, D.R.*
Rapid and reversible knockdown of endogenously tagged endosomal proteins via an optimized HaloPROTAC degrader
ACS Chem. Biol. 2019, 14 (5), 882-892
8. Girardini, M., Maniaci, C., Hughes, S.J., Testa, A., Ciulli, A.*
Cereblon vs VHL: Hijacking E3 Ligases Against Each Other Using PROTACs
Bioorg. Med. Chem. 2019, 27 (12), 2466-2479
7. Roy, M.J., Winkler, S., Hughes, S.J., Whitworth, C., Galant, M., Farnaby, W., Rumpel, K., Ciulli, A.*
SPR-measured dissociation kinetics of PROTAC ternary complexes influence target degradation rate
ACS Chem. Biol. 2019, 14 (3), 361-368
6. Zoppi, V., Hughes, S.J., Maniaci, C., Testa, A., Gmaschitz, T., Wieshofer, C., Koegl, M., Riching, K., Daniels, D.L., Spallarossa, A., and Ciulli, A.*
Iterative design and optimization of initially inactive Proteolysis Targeting Chimeras (PROTACs) identify VZ185 as a potent, fast and selective von Hippel-Lindau (VHL)-based dual degrader probe of BRD9 and BRD7
J. Med. Chem. 2019, 62 (2), 699-726
5. Testa, A., Lucas, X., Castro, G.V., Chan, K.-H., Wright, J.E., Runcie, A.C., Gadd, M.S., Harrison, W.T.A., Ko, E.-J., Fletcher, D., Ciulli, A.*
3-Fluoro-4-hydroxyprolines: Synthesis, conformational analysis and stereoselective recognition by the VHL E3 ubiquitin ligase for targeted protein degradation
J. Am. Chem. Soc. 2018, 140 (29), 9299-9313
4. Chan, K.-H., Zengerle, M., Testa, A., Ciulli, A.*
Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (BET) Degraders Derived from Triazolodiazepine (JQ1) and Tetrahydroquinoline (I-BET726) BET Inhibitor Scaffolds
J. Med. Chem. 2018, 61 (2), 504-513
3. Maniaci, C., Hughes, S.J., Testa, A., Chen, W., Lamont, D.J., Rocha, S., Alessi, D.R., Romeo, R., Ciulli, A.*
Homo-PROTACs: bivalent small-molecule dimerizers of the VHL E3 ubiquitin ligase to induce self-degradation
Nat. Commun. 2017, 8, 830
2. Gadd M.S., Testa A., Lucas X., Chan K.-H., Chen W., Lamont D.J., Zengerle M., Ciulli A.*
Structural basis of PROTAC cooperative recognition for selective protein degradation
Nat. Chem. Biol. 2017, 13 (5), 514-521
1. Zengerle M., Chan K.-H., Ciulli A.*
Selective small molecules induced degradation of the BET bromodomain protein BRD4.
ACS Chem Biol. 2015, 10(8), 1770-1777
Selected references: Reviews
5. Ishida, T., Ciulli, A.*
E3 Ligase Ligands for PROTACs: How They Were Found and How to Discover New Ones
SLAS Discovery, First Published November 3, 2020 DOI: 10.1177/2472555220965528 (Review Article)
4. Zollman, D., Ciulli, A.*
Structural and Biophysical Principles of Degrader Ternary Complexes, in: ‘Protein Degradation with New Chemical Modalities: Successful Strategies in Drug Discovery and Chemical Biology’.
H. Weinmann and C. Crews, Ed., Royal Society of Chemistry, Cambridge, UK, 2020, pp. 14–54.
ISBN: 978-1-78801-686-5. DOI: 10.1039/9781839160691-00014
3. Ciulli, A.*, Farnaby, W.*
Protein degradation for drug discovery. [Editorial]
Drug Discov. Today Technol. 2019 Apr 31: 1-3
2. Maniaci, C., Ciulli, A.*
Bifunctional chemical probes inducing protein-protein interactions
Curr. Opin. Chem. Biol. 2019, 52, 145-156
1. Hughes, S.J., Ciulli, A.*
Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders
Essays in Biochem. 2017, 61, 505-516