Background
Nucleosomes are repeating units of chromatin that are responsible for the primary level of eukaryotic DNA compaction into chromosomes. Each nucleosome consists of an octamer of four histone proteins, H2A, H2B, H3 and H4 and a stretch of double stranded DNA wrapped around the histone core. Nucleosome assembly is highly dynamic which is fundamental in transcriptional regulation and other important biological processes. The crystal structure of the nucleosome core particle has provided unprecedented level of atomic detail of its macromolecular assembly. It has also highlighted how much opportunity there is for molecular recognition, in particular of the histone tails that protrude away from the nucleosome core. Effector or reader proteins bind to histone tails and their posttranslational modifications (PTMs) such as acetylation, methylation and phosphorylation, to recruit components of the transcriptional machinery and chromatin remodeling complexes. PTMs of histone proteins is a crucial signal in the epigenetic regulation of gene transcription. The enzymatic activities that write or erase PTMs on histones appear to be tightly regulated by the recognition of such modifications by evolutionarily-conserved reader domains, however little is known about how specificity is achieved at the molecular level. The importance of these processes to human health and disease and relevant areas of biomedical science and drug discovery are recently being increasingly appreciated, yet a detailed understanding of the molecular and structural determinants of binding specificity and druggability of these reader domains is still lacking.
Research
Our current research is concerned with understanding the molecular recognition of chromatin reader domains and exploit this knowledge to probe their ligandability to small molecules. It can be defined as chemical genetics to probe epigenetics (see our recent review in Curr. Opin. Chem. Biol.).
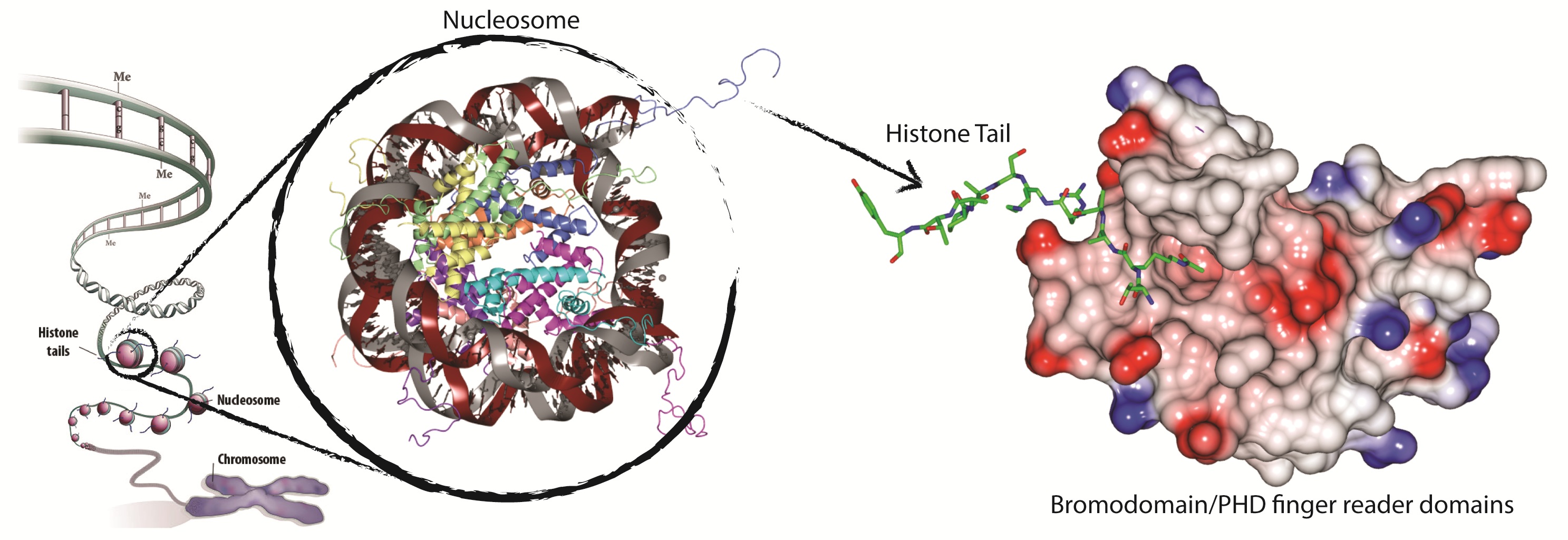
Bromodomains
Bromodomains, found in histone acetyl transferases and other chromatin-associated proteins, are an evolutionarily conserved family of ~110 amino acid modules that specifically recognise acetylated lysines residues. Bromodomains often share highly conserved binding sites amongst family members, therefore achieving target selectivity with chemical probes can be challenging. To address this problem, we are developing novel approaches to engineer ligand selectivity e.g. using chemical genetics. In a paper published in Science we have demonstrated for the first time that it is possible to engineer in a controlled fashion high selectivity of chemical probes targeting the BET subfamily of bromodomains, for example using the landmark triazolodiazepine based BET inhibitors I-BET762/JQ1. Taking this discovery one step further, we have applied the approach inside cells to show that blockade of the first bromodomain of BRD4 is sufficient to displace the protein from chromatin. To achieve this aim, we have developed and optimized various synthetic routes to derivatize and decorate the core aryl-diazepine scaffold of these compounds in highly selective and controlled fashion (Baud et al. J. Med. Chem. 2016) We are now optimizing and further developing the technology to address key biological questions concerning BET protein target validation in health and disease.
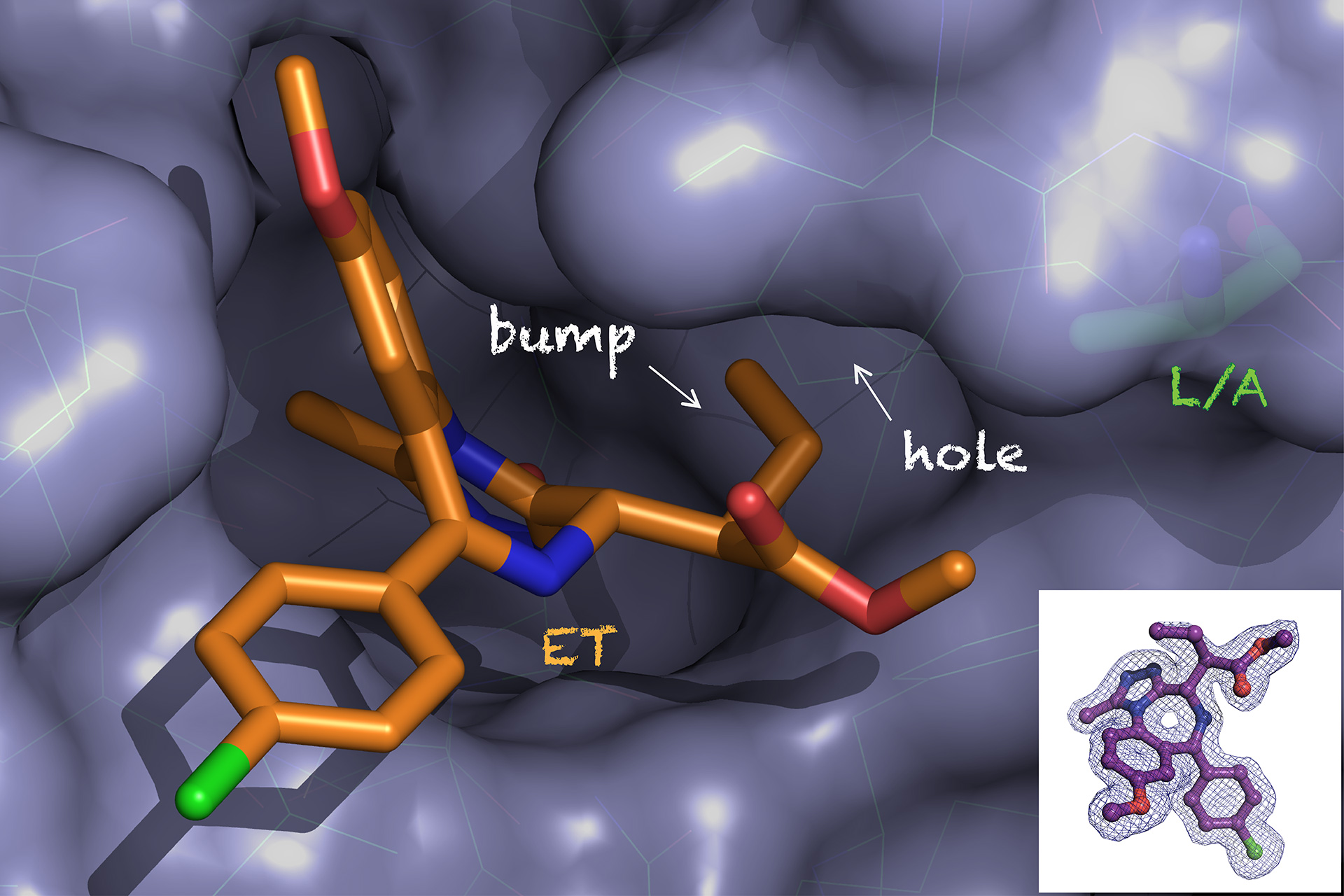
Bump-and-hole chemical genetic approach for allele-selective BET bromodomain inhibition
We are also using fragments to develop novel selective chemical probes of this domain family (see our review in Future Med. Chem.). While BET bromodomains have been widely and successfully targeted with fragments and fragment-derived high-affinity probes, other family member domains have been less readily targeted and their ligandability / tractability to small molecules remains to be addressed. In collaboration with Stefan Knapp (SGC, Oxford) we have demonstrated the application of fragment screening to successfully target BAZ2B, a protein of largely unknown function which bromodomain is predicted to be one of the least druggable in the human phylogenetic tree. We subsequently characterized binding hotspots of the interaction of the BAZ2B bromodomain with its substrate partner, acetylated histone H3K14ac, as revealed by solution NMR spectroscopy and NMR-driven protein-peptide docking, in collaboration with Rolf Boelens, Hans Wienk and Alexandre Bonvin at Utrecht University.
PHD finger domains
Plant homeodomain (PHD) zinc fingers are one of the largest families of epigenetic effectors capable of recognizing modified and unmodified histone tails. PHD zinc fingers are present in a variety of eukaryotic proteins involved in the control of gene transcription and chromatin dynamics. We are interested on interpreting the specific, combinatorial readout of histone patterns by their downstream effectors. One way Nature achieves this is by two or more epigenetic reader domains acting together. The majority of human PHD fingers are indeed found in proteins that contain other histone reader modules, including bromodomains. Structure-function studies on paired reader domains are beginning to shine light on how they could mediate cross-talks between histone PTMs and impart specificity. Yet much work remain to be done to further our still limited understanding of the plasticity of the “histone code” hypothesis. In a recent paper published in Structure, we have combined extensive crystallographic and biophysical studies of the bromodomains and PHD finger domains of BAZ2A (TIP5, a potential drug target involved in epigenetic alterations in prostate cancer) and BAZ2B to show that the bromodomains recognize H4K16ac and H3K14ac, respectively, in both cases with the Kac mark followed by a key basic Arg residue of specific motif: KacXXR – while the PHD finger domain recognize unmodified H3K4. Low-resolution small-angle X-ray scattering (SAXS) data (in collaboration with Dmitri Svergun at Hamburg) for tandem PHD-BRD construct revealed an extended conformation suggesting independent histone tail recognition in trans required for nucleosome recruitment.
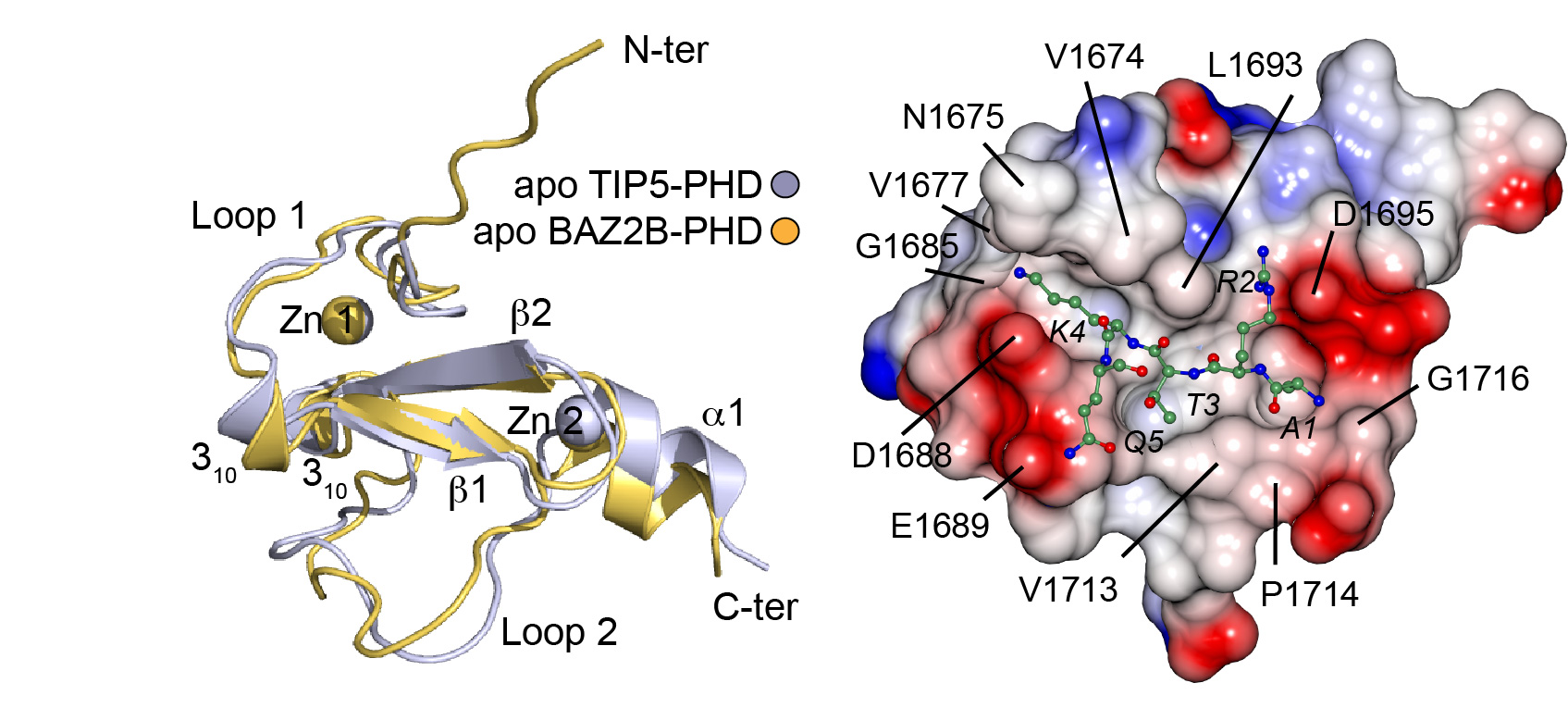
Crystal structures of PHD finger domains with unmodified histone H3 N-terminal tail peptide bound
Selected references: Research articles
9. Bond, A.G., Testa, A., Ciulli, A.*
Stereoselective Synthesis of Allele-Specific BET Inhibitors
Org. Biomol. Chem., 2020, 18, 7533-7539
8. Amato, A., Lucas, X., Bortoluzzi, A., Wright, D., Ciulli, A.*
Targeting ligandable pockets on plant homeodomain (PHD) zinc finger domains by a fragment-based approach
ACS Chem. Biol. 2018, 13 (4), 915-921
7. Runcie, A.C., Zengerle, M., Chan, K.-H., Testa, A., van Beurden, L., Baud, M.G.J., Epemolu, O., Ellis, L.C.J., Read, K.D., Coulthard, V., Brien, A. and Ciulli, A.*
Optimization of a “bump-and-hole” approach to allele-selective BET bromodomain inhibition
Chem. Sci. 2018, 9, 2452-2468
6. Bortoluzzi, A., Amato, A., Lucas, X., Blank, M., Ciulli, A.*
Structural Basis of Molecular Recognition of Helical Histone H3 Tail by PHD Finger Domains.
Biochem. J. 2017, 474(10), 1633-1651
5. Baud, M.G., Lin-Shiao, E., Zengerle, M., Tallant, C., Ciulli, A.*
New Synthetic Routes to Triazolo-Benzodiazepine Analogues: Expanding the Scope of the Bump-and-Hole Approach for Selective BET Bromodomain Inhibition.
J. Med. Chem. 2016, 59(4), 1492-1500
4. Tallant C., Valentini E., Fedorov O., Overvoorde L., Ferguson F.M., Filippakopoulos P., Svergun D.I., Knapp S., Ciulli A.*
Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure 2015, 23(1), 80-92
3. Baud M.G., Lin-Shiao E., Cardote T., Tallant C., Pschibul A., Chan K.H., Zengerle M., Garcia J.R., Kwan T.T., Ferguson F.M., Ciulli A.*
A bump-and-hole approach to engineer controlled selectivity of BET bromodomain chemical probes.
Science 2014, 346(6209), 638-41
2. Ferguson F.M., Dias D.M., Rodrigues J.P., Wienk H., Boelens R., Bonvin A.M., Abell C., Ciulli A.*
Binding hotspots of BAZ2B bromodomain: Histone interaction revealed by solution NMR driven docking.
Biochemistry 2014, 53(42), 6706-16
1. Ferguson, F.M., Fedorov, O., Chaikuad, A., Philpott, M., Muniz, J., Felletar, I., von Delft, F., Heightman, T.D., Knapp, S., Abell, C., Ciulli, A.*
Targeting Low-Druggability Bromodomains: Fragment Based Screening and Inhibitor Design Against the BAZ2B Bromodomain.
J. Med. Chem. 2013, 56(24), 10183-10187
Selected referenced: Reviews
3. Cipriano, A., Sbardella, G.,* Ciulli, A.*
Targeting epigenetic reader domains by chemical biology
Curr. Opin. Chem. Biol. 2020, 57, 82-94
2. Runcie, A.C., Chan, K.-H., Zengerle, M., Ciulli, A.*
Chemical genetics approaches for selective intervention in epigenetics.
Curr. Opin. Chem. Biol. 2016, 33, 186-194
1. Galdeano, C., Ciulli, A.*
Selectivity on-target of bromodomain chemical probes by structure-guided medicinal chemistry and chemical biology
Future Med. Chem. 2016, 8(13), 1655-1680