Chemical intervention within our biological systems of interest requires small molecule modulation of protein-protein interactions (PPIs). Combining biophysical techniques (TS, FP, SPR, NMR, ITC, X-ray crystallography amongst others) for screening, hit validation and structural characterisation, we are aiming at discovering and characterising binding hotspots at protein-protein interfaces, with the ultimate goal to develop and deliver novel high quality small molecule ligands and probes for biology. In this context, biophysical fragment screening is a powerful way for the discovery of new ligands to target known as well as novel binding sites on protein surfaces that can rapidly lead to high-quality chemical probes modulators of PPIs. The structural diversity of fragments allows for efficient chemical space evaluation, and potentially provide a number of invaluable starting points for further structure-based rational design and lead optimisation via iterative cycles of synthesis and evaluation of binding affinities.
A major challenge is that the low ligand efficiencies (LEs) typically associated with small molecule binding at protein surfaces and PPIs raise the barrier to the detection of bona fide low-affinity fragment hits. One way to approach this challenge would be to screen focused libraries of slightly larger compounds and ad-hoc pysicochemical property compared to routinely used fragment-like compounds. Our lab’s work, published in Chemistry & Biology, suggests that the most fruitful size of fragments to screen against PPI targets should be somewhat larger than commonly used for more druggable targets, pushing the limits posed by the Rule of Three (Ro3), and could include “privileged” scaffolds such as aromatic-aminoacid hybrid compounds. An alternative approach toward this problem would be to increase the sensitivity of binding detection in a fragment screen. In a second paper from our laboratory, subsequently published in ACS Med. Chem. Lett., we interrogate the detection limits of weak fragment binding at a known druggable PPI by ligand-based NMR spectroscopy techniques. We show that screening at higher receptor and ligand concentrations opens the possibility of rescuing genuine Ro3-compliant binders as true hits. This revision on the experimental NMR conditions will deliver more sensitive and robust set-ups for NMR fragment screening, which in turn will enable a more reliable assessment of the druggability and tractability to small molecules of PPI targets. Amongst key challenges posed by applying fragment-based drug discovery to targeting large multi-protein complexes such as those that most commonly function in crucial cell signaling and regulatory pathways are those that relate specifically with enabling structural biology -driven structure-based design projects against these targets, using e.g. protein-observed NMR spectroscopy (Dias & Ciulli, Prog. Biophys. Mol. Biol. 2014) and ligand soaking or co-crystallization by X-ray crystallography.
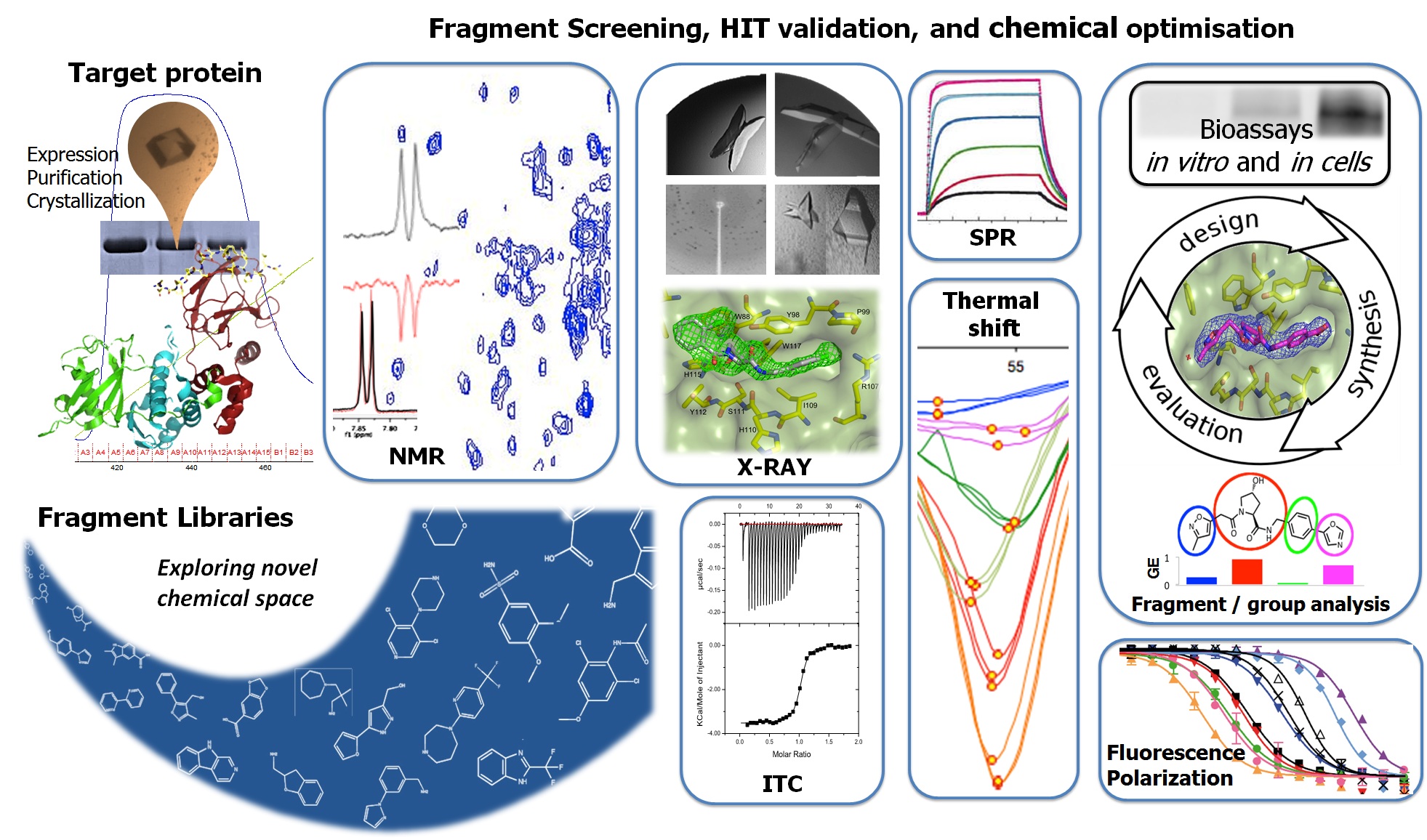
We are also engaged successfully with collaborators to apply fragment screening to identify starting hits against challenging targets of interest. For example, collaboration with Professor Barbara Imperiali (MIT) have helped targeting sugar-binding sites enzymes involved in targeting bacillosamine biosynthesis in bacterial pathogens – and specifically using fragment-based approaches to develop inhibitors against a bacterial amino-sugar acetyltransferase from Campylobacter jejuni (De Schutter et al. J. Med. Chem. 2017).
Selected references: Research articles
11. Castro, G.V, Ciulli, A.*
Spy vs. spy: selecting the best reporter for 19F NMR competition experiments
Chem. Commun. 2019, 55 (10), 1482-1485
10. Lucas, X., Van Molle, I., Ciulli, A.*
Surface probing by fragment-based screening and computational methods identifies ligandable pockets on the von Hippel-Lindau (VHL) E3 ubiquitin ligase
J. Med. Chem. 2018, 61 (16), 7387-7393
9. Amato, A., Lucas, X., Bortoluzzi, A., Wright, D., Ciulli, A.*
Targeting ligandable pockets on plant homeodomain (PHD) zinc finger domains by a fragment-based approach
ACS Chem. Biol. 2018, 13 (4), 915-921
8. Morreale, F.E., Testa, A., Chaugule, V.K., Bortoluzzi, A., Ciulli, A.*, Walden, H.*
Mind the metal: a fragment library-derived zinc impurity binds the E2 ubiquitin-conjugating enzyme Ube2T and induces structural rearrangements
J. Med. Chem. 2017, 60 (19), 8183-8191
7. Morreale, F.E., Bortoluzzi, A., Chaugule, V.K., Arkinson, C., Walden, H.*, Ciulli, A.*
Allosteric targeting of the Fanconi anemia ubiquitin-conjugating enzyme Ube2T by fragment screening
J. Med. Chem. 2017, 60 (9), 4093-4098
6. Ryan, A., Polycarpou, E., Lack, N.A., Evangelopoulos, D., Sieg, C., Halman, A., Bhakta, S., Eleftheriadou, O., McHugh, T.D., Keany, S., Lowe, E.D., Ballet, R., Abuhammad, A., Jacobs, W.R. Jr, Ciulli, A., Sim, E.
Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach
Br. J. Pharmacol. 2017, 174 (14), 2209-2224
5. De Schutter, J.W., Morrison, J.P., Morrison, M.J., Ciulli, A., Imperiali, B.
Targeting Bacillosamine Biosynthesis in Bacterial Pathogens: Development of Inhibitors to a Bacterial Amino-Sugar Acetyltransferase from Campylobacter jejuni
J. Med. Chem. 2017, 60 (5), 2099-2118
4. Dias, D.M., Van Molle, I., Baud, M.G.J., Galdeano, C., Geraldes, C.F.G.C., Ciulli, A.*
Is NMR Fragment Screening Fine-Tuned to Assess Druggability of Protein-Protein Interactions?
ACS Med. Chem. Lett. 2014, 5(1), 23-28
3. Ferguson, F.M., Fedorov, O., Chaikuad, A., Philpott, M., Muniz, J., Felletar, I., von Delft, F., Heightman, T.D., Knapp, S., Abell, C., Ciulli, A.*
Targeting Low-Druggability Bromodomains: Fragment Based Screening and Inhibitor Design Against the BAZ2B Bromodomain.
J. Med. Chem. 2013, 56(24), 10183-10187
2. Silvestre, H.L., Blundell, T.L., Abell, C., Ciulli, A.*
Integrated Biophysical Approach to Fragment Screening and Validation for Fragment-Based Lead Discovery.
Proc. Natl. Acad. Sci. U.S.A. 2013, 110(32), 12984-12989
1. Van Molle, I., Thomann, A., Buckley, D.L., So, E.C., Lang, S., Crews, C.M., Ciulli, A.*
Dissecting Fragment-Based Lead Discovery at the von-Hippel Lindau Protein : Hypoxia Inducible Factor 1α Protein-Protein Interface.
Chemistry & Biology 2012, 19, 1300–1312
Selected articles: Reviews
4. Bortoluzzi, A., Ciulli, A.*
Protein-Based NMR Methods Applied to Drug Discovery, in: ‘Applied Biophysics in Drug Discovery’.
D. Huddler and E. R. Zartler, Ed., John Wiley & Sons. 2017, pp. 153-173. ISBN: 978-1-119-09948-2
DOI: 10.1002/9781119099512.ch9
3. Dias, D.M., Ciulli, A.*
NMR approaches in structure-based lead discovery: recent developments and new frontiers for targeting multi-protein complexes.
Prog Biophys Mol Biol. 2014, 116(2-3), 101-12
2. Ciulli, A.*
Biophysical Screening for the Discovery of Small-Molecule Ligands.
In ‘Protein-Ligand Interactions – Methods and Applications’, 2nd Ed., T. Daviter and M. Williams, Ed., Springer Protocols, Humana Press. ISBN: 978-1-62703-397-8
Methods Mol. Biol. 2013, 1008, 357–388
1. Śledź, P., Abell, C., Ciulli, A.*
Ligand‐Observed NMR in Fragment‐Based Approaches, in: ‘NMR of Biomolecules: Towards Mechanistic Systems Biology’.
I. Bertini, K. McGreevy and G. Parigi, Ed., Wiley, 2012, pp. 265–280. ISBN: 978-3-527-32850-5
DOI: 10.1002/9783527644506.ch15