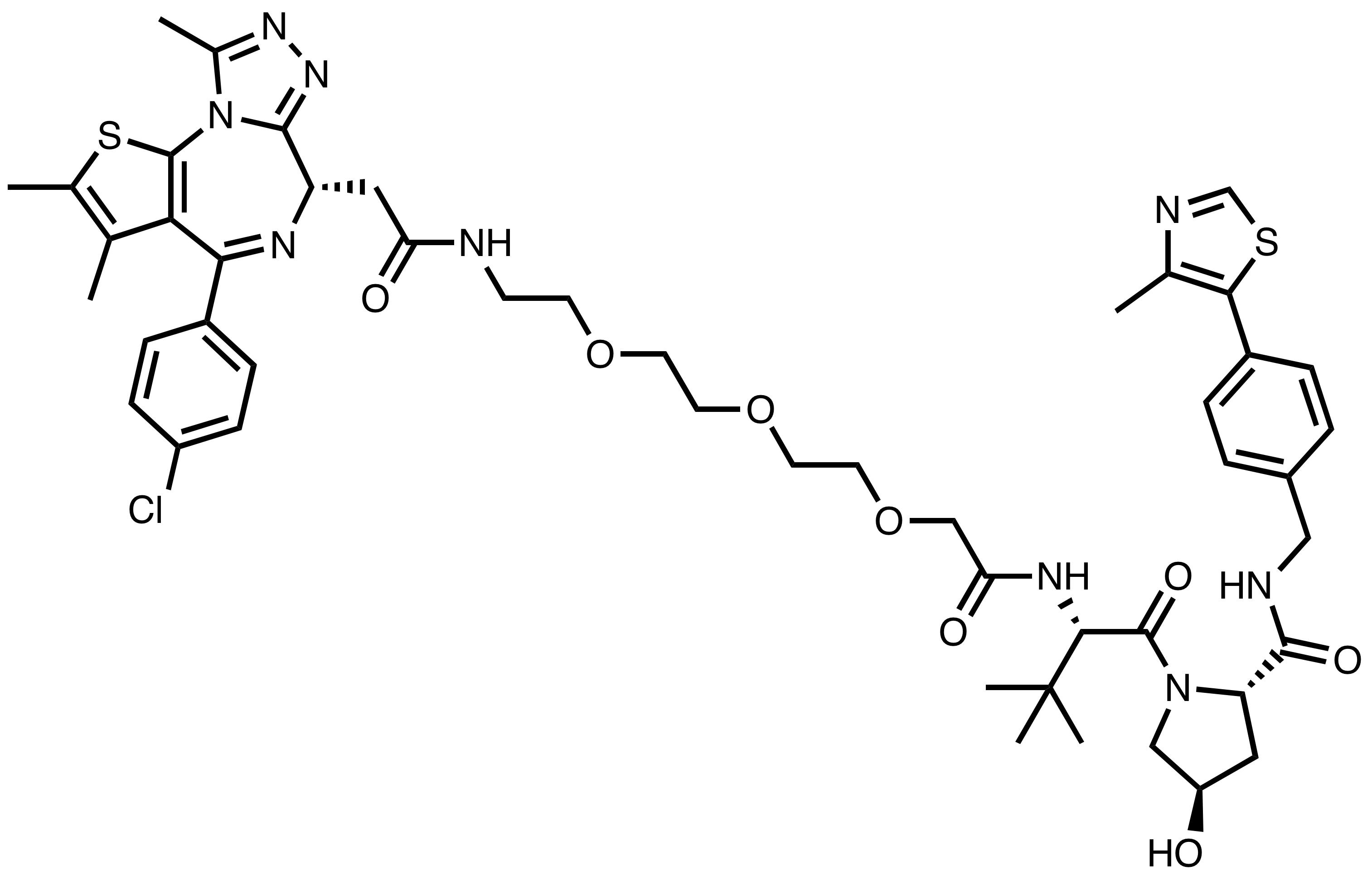
MZ1
Protein Target(s) Name: BET proteins BRD4, BRD3, BRD2
Mechanism of Action: PROTAC degrader
Description: VH032 (VHL) based and (+)-JQ1 based PROTAC that degrades BET proteins in cells and in vivo. MZ1 exhibits preferential degradation of BRD4 at nM concentrations (1).
Chemical Name: (2S,4R)-1-((S)-2-(tert-butyl)-17-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-4,16-dioxo-6,9,12-trioxa-3,15-diazaheptadecanoyl)- 4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide
CAS Number: 1797406-69-9
In vitro pharmacology*: MZ1 reduces BET protein levels in human cells: Brd4 pDC50 | Dmax (%) in HeLa cells (24 h) = 8.6 | 100
MZ1 shows antiproliferative and Myc-suppression activity in AML MV4;11 and HL60 cells: pEC50 in MV4;11 | HL-60 cells (48 h) = 7.6 | 6.7
Data from ref. (3)
*DC50: concentration in molar causing 50% reduction of protein level relative to vehicle control treatment.
Dmax: maximum reduction of protein level relative to vehicle control treatment.
EC50: effective concentration in molar causing 50% reduction of cell viability relative to vehicle control treatment.
Biophysical binding data: ITC binary Kd (Brd4-BD2) = 15 nM; ITC binary Kd (VHL) = 66 nM; ITC ternary Kd (VHL, in the presence of Brd4-BD2) = 3.7 nM; cooperativity (alpha) = 18. Ternary complex stability DeltaG = -22.2 kcal/mol. Data from ref. (2)
Ternary complex VHL:MZ1:Brd4-BD2 t1/2 (SPR) = 130 s. FP ternary Kd (VHL, in the presence of Brd4-BD2) = 1.3 nM. Data from ref. (4)
In vivo PK data: MZ1 is suitable for a parenteral administration (i.v., i.p. or s.c.). The compound shows high clearance in rats and low clearance in mice. High AUC levels can be obtained, when the compound is administered subcutaneously using a 25% HP-ß-CD formulation. Because of the high Caco2 efflux ratio, the oral exposure is very low. Full detail of the in vivo PK data are available from OpnMe.com
Crystal Structure: ternary complex EloBC-VHL : MZ1 : Brd4(BD2). PDB entry code: 5T35 (2)
Negative control: cis MZ1, CAS number: 1797406-72-4, available from Tocris
Primary References:
- Zengerle et al. (2015) Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS. Chem. Biol. 10, 1770. DOI: 10.1021/acschembio.5b00216; PMID: 26035625
- Gadd et al. (2017) Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514. DOI: 10.1038/nchembio.2329; PMID: 28288108
- Chan et al. (2018) Impact of Target Warhead and Linkage Vector on Inducing Protein Degradation: Comparison of Bromodomain and Extra-Terminal (BET) Degraders Derived From Triazolodiazepine (JQ1) and Tetrahydroquinoline (I-BET726) BET Inhibitor Scaffolds. J. Med. Chem. 61, 504. DOI: 10.1021/acs.jmedchem.6b01912; PMID: 28595007
- Roy et al. (2019) SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem. Biol. 14, 361. DOI: 10.1021/acschembio.9b00092; PMID: 30721025
Articles that have used MZ1:
- 2017 Fernandez-Alonso EMBO reports http://dx.doi.org/10.15252/embr.201643534
- 2017 Gadd Nat Chem Biol http://dx.doi.org/10.1038/nchembio.2329
- 2017 Chan J Med Chem http://dx.doi.org/10.1021/acs.jmedchem.6b01912
- 2017 Wurz J Med Chem http://dx.doi.org/10.1021/acs.jmedchem.6b01781
- 2018 Sansam Genes & Dev http://dx.doi.org/10.1101/gad.306464.117
- 2018 Leonard Cancer Research http://dx.doi.org/10.1158/0008-5472.CAN-18-0459
- 2018 Nowak Nat Chem Biol https://doi.org/10.1038/s41589-018-0055-y
- 2018 Testa J Am Chem Soc http://dx.doi.org/10.1021/jacs.8b05807
- 2018 Riching ACS Chem Biol http://dx.doi.org/10.1021/acschembio.8b00692
- 2018 Aresu Haematologica http://dx.doi.org/10.3324/haematol.2018.207027
- 2018 Jensen Front Immunol https://dx.doi.org/10.3389%2Ffimmu.2018.02697
- 2019 Roy ACS Chem Biol http://dx.doi.org/10.1021/acschembio.9b00092
- 2018 Spradlin Nat Chem Biol http://dx.doi.org/10.1038/s41589-019-0304-8
- 2018 Ward ACS Chem Biol http://dx.doi.org/10.1021/acschembio.8b01083
- 2019 Piya J Clin Invest https://doi.org/10.1172/JCI120654
- 2019 Tsujikawa Clin Epigenetics https://doi.org/10.1186/s13148-019-0696-z
- 2019 Lim Haematologica https://doi.org/10.3324/haematol.2018.201483
- 2019 Andrieu Cancer Lett https://doi.org/10.1016/j.canlet.2019.08.013
- 2019 Noblejas-López J Exp Clin Cancer Res https://doi.org/10.1186/s13046-019-1387-5
- 2019 Ottis ACS Chem Biol http://dx.doi.org/10.1021/acschembio.9b00525
- 2019 Pillow ChemMedChem https://doi.org/10.1002/cmdc.201900497
- 2019 Otto Neoplasia https://doi.org/10.1016/j.neo.2019.10.003
- 2019 Shafran Mol Cancer Res https://doi.org/10.1158/1541-7786.mcr-18-1279
- 2020 Jiang Gastroenterology https://doi.org/10.1053/j.gastro.2020.06.050
- 2020 Kaji Sci Reports https://doi.org/10.1038/s41598-020-59966-5
- 2020 Dubois Mol Syst Biol http://doi.org/10.15252/msb.20199156
- 2020 Testa Angew Chem Intl Ed https://doi.org/10.1002/anie.201914396
- 2020 Foley ACS Chem Biol https://doi.org/10.1021/acschembio.9b00972
- 2020 Rosencrance Mol Cell https://doi.org/10.1016/j.molcel.2020.03.018
- 2020 Kounde Chem Commun https://doi.org/10.1039/D0CC00523A
- 2020 Ermondi Drug Discov Today https://doi.org/10.1016/j.drudis.2020.06.015
- 2020 Beveridge ACS Cent Sci https://doi.org/10.1021/acscentsci.0c00049
- 2020 Shafran Prostate Cancer Prostatic Dis https://doi.org/10.1038/s41391-020-0246-y
- 2020 Klein ACS Med Chem Lett https://doi.org/10.1021/acsmedchemlett.0c00265
- 2020 Cimas Pharmaceutics https://doi.org/10.3390/pharmaceutics12100986
- 2020 Lin Bioconjugate Chem https://doi.org/10.1021/acs.bioconjchem.0c00507
- 2020 Li Front Oncol https://doi.org/10.3389/fonc.2020.574525
- 2021 Shirasaki Cell Reports https://doi.org/10.1016/j.celrep.2020.108532
- 2021 Alpsoy Cancer Res https://doi.org/10.1158/0008-5472.CAN-20-1417
- 2021 Kaiho-Soma Mol Cell https://doi.org/10.1016/j.molcel.2021.01.023
- 2021 Dragovich J Med Chem https://doi.org/10.1021/acs.jmedchem.0c01845
- 2021 Dragovich J Med Chem https://doi.org/10.1021/acs.jmedchem.0c01846
- 2021 Wang Cell Death Differ https://doi.org/10.1038/s41418-021-00751-w
- 2021 Noblejas-López J Exp Clin Cancer Res https://doi.org/10.1186/s13046-021-01907-9
- 2021 Pietrobono Oncogene https://doi.org/10.1038/s41388-021-01783-9
- 2021 Song Cell Chem Biol https://doi.org/10.1016/j.chembiol.2021.05.005
- 2021 Wang Nat Commun https://doi.org/10.1038/s41467-021-24687-4
- 2021 Massafra J Immunol https://doi.org/10.4049/jimmunol.2000252
- 2021 Bhola Head & Neck https://doi.org/10.1002/hed.26827
- 2021 Castro RSC Med Chem https://doi.org/10.1039/D1MD00215E
- 2021 Tong Oncogene https://doi.org/10.1038/s41388-021-02041-8
- 2021 Bond J Med Chem https://doi.org/10.1021/acs.jmedchem.1c01532
-
2021 Divakaran ACS Med Chem Lett https://doi.org/10.1021/acsmedchemlett.2c00300; ChemRxiv https://doi.org/10.33774/chemrxiv-2021-zvq4t
-
2021 Wang Doctoral thesis http://hdl.handle.net/21.11130/00-1735-0000-0005-153B-2
-
2021 Imaide Nat Chem Biol https://doi.org/10.1038/s41589-021-00878-4
-
2021 Jafari Science Signal. https://doi.org/10.1126/scisignal.abj2807
-
2021 Weng J Med Chem https://doi.org/10.1021/acs.jmedchem.1c01576
-
2021 Klein J Med Chem https://doi.org/10.1021/acs.jmedchem.1c01496
-
2021 Tarantelli Explor. Target Antitumor Ther. https://doi.org/10.37349/etat.2021.00065
-
2022 Müller Clin Epigenet https://doi.org/10.1186/s13148-021-01223-1
-
2022 Weerakoon J Chem Inf Model https://doi.org/10.1021/acs.jcim.1c01036
- 2022 Luo iScience https://doi.org/10.1016/j.isci.2022.103985
- 2023 Hanzl Nat Chem Biol https://doi.org/10.1038/s41589-022-01177-2; 2022 BioRxiv https://doi.org/10.1101/2022.04.14.488316
- 2022 García Jiménez J Med Chem https://doi.org/10.1021/acs.jmedchem.2c00201
- 2022 Trapotsi ACS Chem Biol https://doi.org/10.1021/acschembio.2c00076
- 2022 Mason BioRxiv https://doi.org/10.1101/2022.10.13.512184
- 2022 Bhela J Med Chem https://doi.org/10.1021/acs.jmedchem.2c01218
- 2023 Toriki ACS Cent Sci https://doi.org/10.1021/acscentsci.2c01317; 2022 BioRxiv https://doi.org/10.1101/2022.11.04.512693
- 2022 Lou Science https://doi.org/10.1126/science.abl5829
- 2023 Bashore Nat Chem Biol https://doi.org/10.1038/s41589-022-01218-w
- 2023 Shergalis ACS Chem Biol https://doi.org/10.1021/acschembio.2c00747
- 2023 Hsia BioRxiv https://doi.org/10.1101/2023.02.14.528511
- 2023 Li BioRxiv https://doi.org/10.1101/2023.02.14.528208
- 2023 Singh Sci Adv https://doi.org/10.1126/sciadv.ade3876
- 2023 Steinebach ChemRxiv https://doi.org/10.26434/chemrxiv-2023-zshqp
- 2023 Bordi Digital Discovery https://doi.org/10.1039/D3DD00027C
- 2023 Tong Nature https://doi.org/10.1038/s41586-023-06091-8
- 2023 Bagka Nat Commun https://doi.org/10.1038/s41467-023-39657-1
- 2023 Plesniak ChemRxiv https://doi.org/10.26434/chemrxiv-2023-dz9l7
- 2023 Hales Chemistry https://doi.org/10.1002/chem.202301975
More information
More information on our chemical probes can be found from the Chemical Probes portal, and from commercial vendors such as Tocris Bio-Techne.