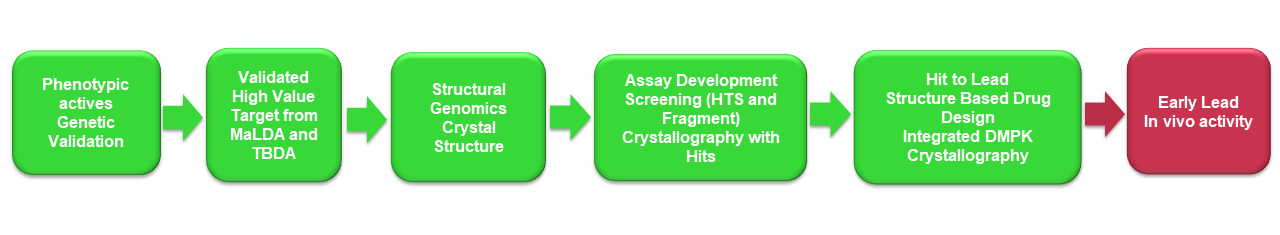
Our Approach to speed up Malaria and Tuberculosis drug discovery
dSDDC primary aim is to develop compounds to “Early Lead” status, with Proof-of-Concept in animal models of infection for Malaria and Tuberculosis
We achieve this running structure-guided drug discovery program on thoroughly validated targets for these diseases. Target validation is typically through association of a phenotypically (whole cell) active compound with the molecular target. These targets are then paired with crystal structures giving rise to structure-guided hit to lead projects. The Drug Discovery Unit (DDU-Dundee) based at the University of Dundee, is responsible for the medicinal chemistry development of the SDDC projects devoted to finding potential new start points for antimalarial and antituberculosis drug discovery. The collaborating structural genomic groups, including the structural biology laboratories at Seattle Structural Genomics Center for Infectious Disease (SSGCID-Seattle), Center for Structural Genomics of Infectious Disease (CSGID-Chicago), the University of Campinas (UNICAMP-Campinas) are the key collaborators for protein production, assay development, structural biology. The structural genomic centres generate high resolution Xray structures with phenotypically active inhibitors showing how the molecules bind to the target protein. This information is very important to guide modifications to improve their activity.
Combining the value of structure-guided Medicinal Chemistry with the confidence that the starting hit achieves its disease-relevant effect through interaction with the validated target, is an especially powerful approach.
The coalition will also emphasize the incorporation of new technologies into discovery and optimization process, including, but not limited to, fragment-based screening, machine learning/artificial intelligence and high throughput plate-based chemistry.
Critieria for target selection
The majority of validated targets will come from MALDA and TBDA.
Both MALDA and the TBDA have working groups for prioritizing targets. These are broadly based on the following criteria (1, 2)
- Essentiality (genetic and chemical)
- Druggability
- TPP (target product profile)/TCP (target candidate profile) fit
- Potential for selectivity
- Low resistance potential
- Assay feasibility
- Structurally enabled
The target prioritization groups in MALDA and the TBDA are periodically reviewing targets – as more data becomes available so targets may move up or down the prioritization list. However, for the purposes of the SDDC, we focus on those targets that are highly validated. We liaise closely with those groups doing target prioritization; the DDU is closely involved in these target prioritization assessment teams in both MALDA and TBDA.
References
(1) Prioritization of Molecular Targets for Antimalarial Drug Discovery
DOI: 10.1021/acsinfecdis.1c00322
(2) Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis
Barbara Bosch, Michael A. DeJesus, Nicholas C. Poulton, Wenzhu Zhang, Curtis A. Engelhart, Anisha Zaveri, Sophie Lavalette, Nadine Ruecker, Carolina Trujillo, Joshua B. Wallach, Shuqi Li, Sabine Ehrt, Brian T. Chait, Dirk Schnappinger, Jeremy M. Rock
Cell, Volume 184, Issue 17, 2021, 4579-4592.e24
ISSN 0092-8674
https://doi.org/10.1016/j.cell.2021.06.033.University of