Clouds in the sky get their white colour from the scattering of light. This is a well-understood phenomenon, yet notoriously difficult to compute. Deep-tissue microscopy relies on controlling light scattering within biological tissue, so we take a particular interest in this issue. By mapping Maxwell’s equations onto the structure of a neural network, we were able to scale up light wave scattering calculations on the cloud, more specifically, the Google Colab cloud.
We integrated this into the open source MacroMax EM solver, ideal for computing coherent scattering in highly-heterogeneous materials such as biological tissue. It is straighforward to install and use with Python. Well within 10 minutes, light wave scattering can now be computed throughout a complex 3D structure with all 3 sides >100μm!
Read more about how this works in our recently published paper doi: 10.34133/icomputing.0098.
 Together with K. Dholakia I authored a book chapter “Shaped Beams for Light Sheet Imaging and Optical Manipulation” in the book “Wavefront Shaping for Biomedical Imaging” (online ISBN:
Together with K. Dholakia I authored a book chapter “Shaped Beams for Light Sheet Imaging and Optical Manipulation” in the book “Wavefront Shaping for Biomedical Imaging” (online ISBN: 
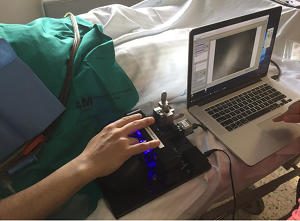 Our work on automated detection of neutropenia is published in the American Journal for Hematology! The blood flow in the capillaries is imaged through the nail-fold skin. Machine learning techniques are applied to detect the location of the capillaries in the image, and spatio-temporal correlations are analysed per capillary. Read more here:
Our work on automated detection of neutropenia is published in the American Journal for Hematology! The blood flow in the capillaries is imaged through the nail-fold skin. Machine learning techniques are applied to detect the location of the capillaries in the image, and spatio-temporal correlations are analysed per capillary. Read more here: 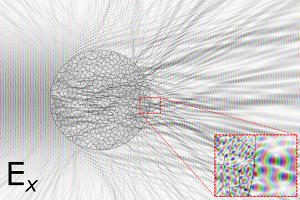 Biological samples, often the subject of optical microscopy, tend to be rather heterogeneous. This affects the propagation of the electromagnetic field of light. While the Maxwell’s laws underlying the propagation of electromagnetic waves in such tissue are well-understood; accurate numerical calculation does not scale well. Even the sub-millimeter-sized sample areas in microscopy pose significant challenges. Recently this changed. Osnabrugge et al. proposed a
Biological samples, often the subject of optical microscopy, tend to be rather heterogeneous. This affects the propagation of the electromagnetic field of light. While the Maxwell’s laws underlying the propagation of electromagnetic waves in such tissue are well-understood; accurate numerical calculation does not scale well. Even the sub-millimeter-sized sample areas in microscopy pose significant challenges. Recently this changed. Osnabrugge et al. proposed a