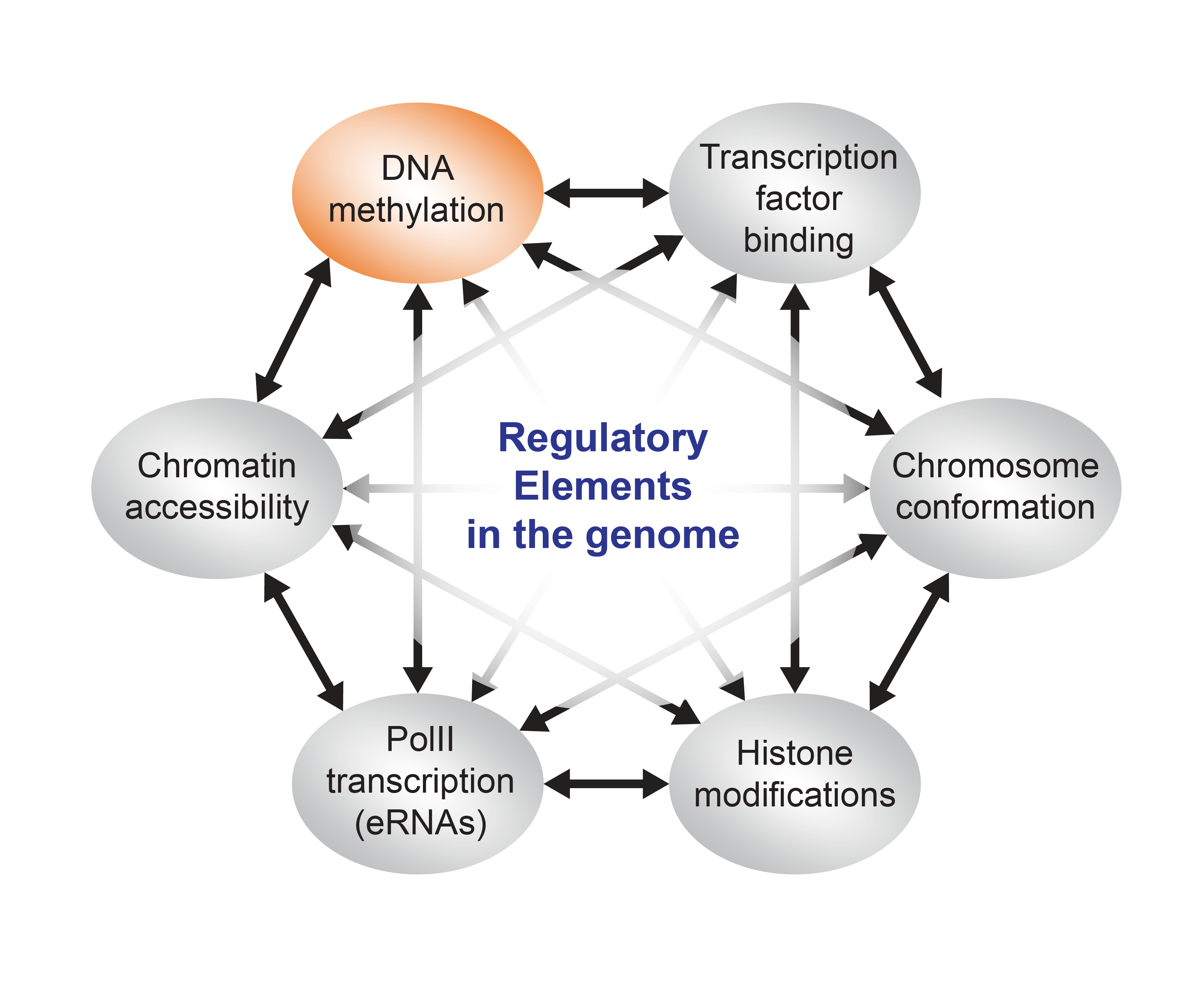
DNA methylation is a chemical modification found on a majority of cytosines in CG dinucleotides in the human genome. This reversible epigenetic mark has been implicated in many important biological processes (e.g. genomic imprinting, X-chromosome inactivation, silencing of transposable elements etc. etc.), however the role of DNA methylation in regulation of gene expression is still a matter of debate.
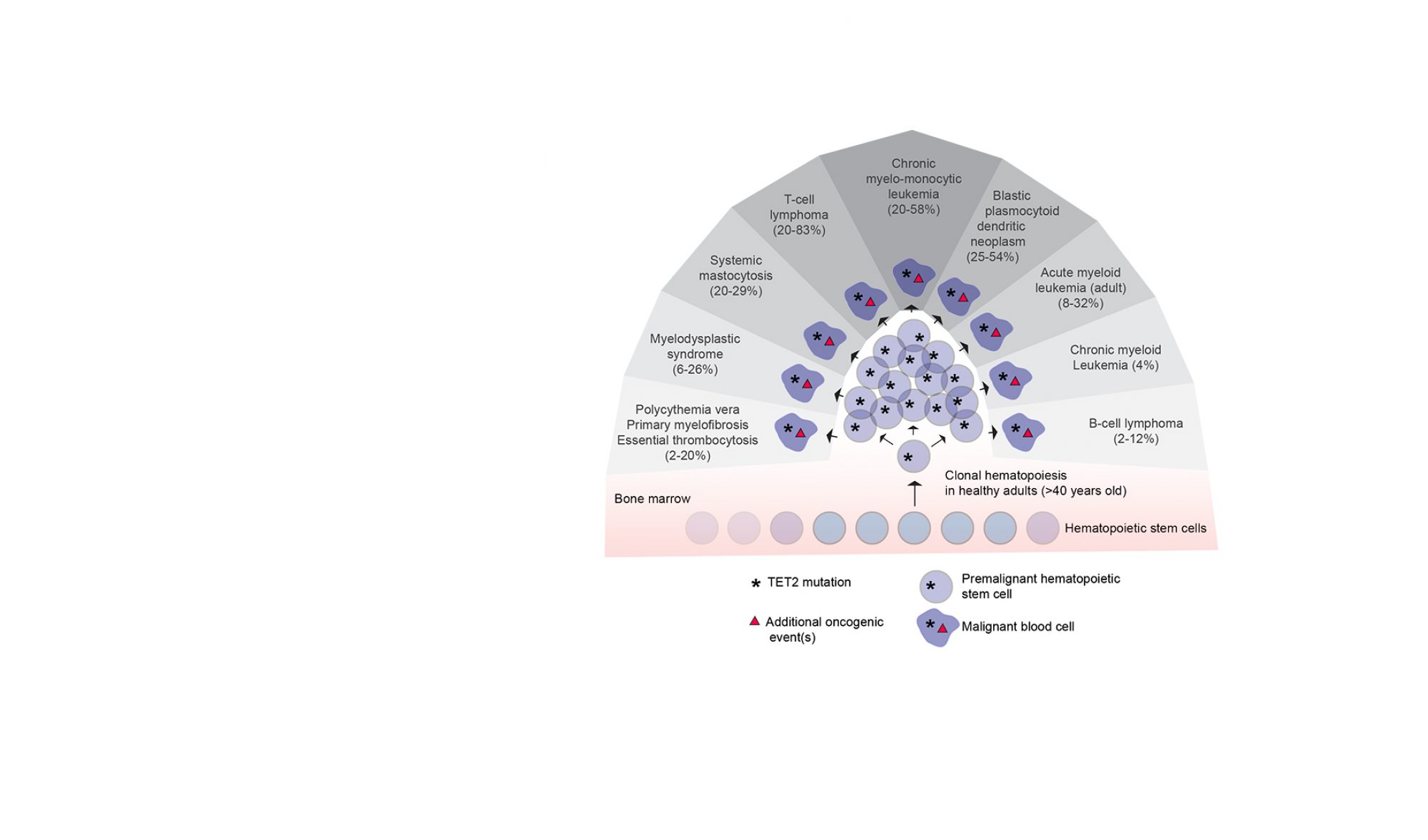
Leukemias are a group of diseases arising from the blood-forming cells in the body. The gene encoding TET2; an “epigenetic eraser” important for removal of methylation of DNA, is often mutated in blood cancers. However, the downstream molecular events following the occurrence of a mutation are poorly understood. We and others have shown that blood cells lacking TET2 are particularly prone to accumulate DNA methylation at specific regulatory elements in the genome, often defined as enhancers, which are important to control gene expression.
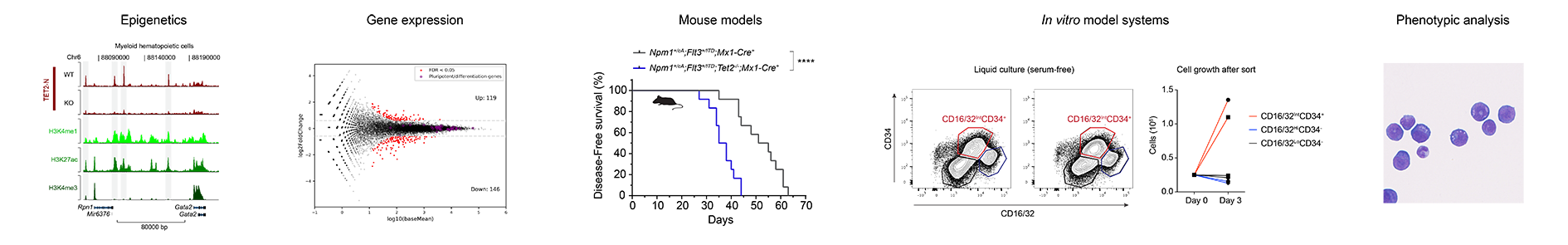
The aim of our work is to dissect the mechanisms by which the proteins regulating the DNA methylation landscape affects chromatin structure and gene expression patterns in hematopoietic cells. To do this, we utilize CRISPR genome editing in embryonic stem cells, mouse and human hematopoietic cell lines and complex mouse models of leukemia and combine these with the application of new biochemical, genomics and proteomics techniques. The biological insights obtained may provide the basis for developing targeted therapies for blood cancers with mutations in the DNA methylation machinery.
Current topics in the lab are:
- Identification of TET2 recruiters and interactors and their role in TET2 function in hematopoietic cells
- Defining an instructive role of enhancer DNA methylation in regulation of gene expression
- Application of third-generation sequencing technologies to DNA methylation analysis
- The role of TET2 in Blastic Plasmacytoid Dendritic Cell Neoplasm