Molecular mechanisms controlling asymmetric stem cell division
We are interested in identifying aspects of cellular asymmetry and the control of cell polarity. We use neural stem cells, called neuroblasts, of the developing Drosophila brain as a model system. At mitosis neuroblast polarity is most prominent at the cell cortex where different protein complex localise to the apical and basal cell pole defining the neuroblast polarity axis. We are currently investigating how the different domains are established.
The division is asymmetric as one daughter cell will self-renew as a neuroblast, while the other daughter cell will enter a different developmental programme and differentiate. In this way, the population fo stem cells is controlled, while multiple neurons and glia cells are generated.
The Video shows a neuroblast in culture in metaphase. The apical cortex (green) is labled with Par-3/Bazooka GFP and the basal cortex is decorated with mCherry-Miranda, an adaptor protein carrying fate determinants such as Prospero.
Mechanisms orienting stem cell division?
The orientation of cell division is an important regulatory mechanism that controls what microenvironment cells are exposed to and what neighbouring cells they will be in contact with. This process is important for cell fate specification and also contributes to the morphogenesis of tissues and organs. Neuroblasts in the developing Drosophila brain keep their division axis remarkably constant, yet the mechanism that control this process are not clear, nor is it understood, why these cells keep their orientation constant
In neuroblast, the orientation of division is defined by the orientation of the apical basal polarity axis. This axis is established de novo at each mitosis, raising the question, how the orientation of the axis is stably maintained between consecutive divisions. The neuroblasts system is therefore also ideally suited to study the spatial cues that define the orientation of cell polarity.
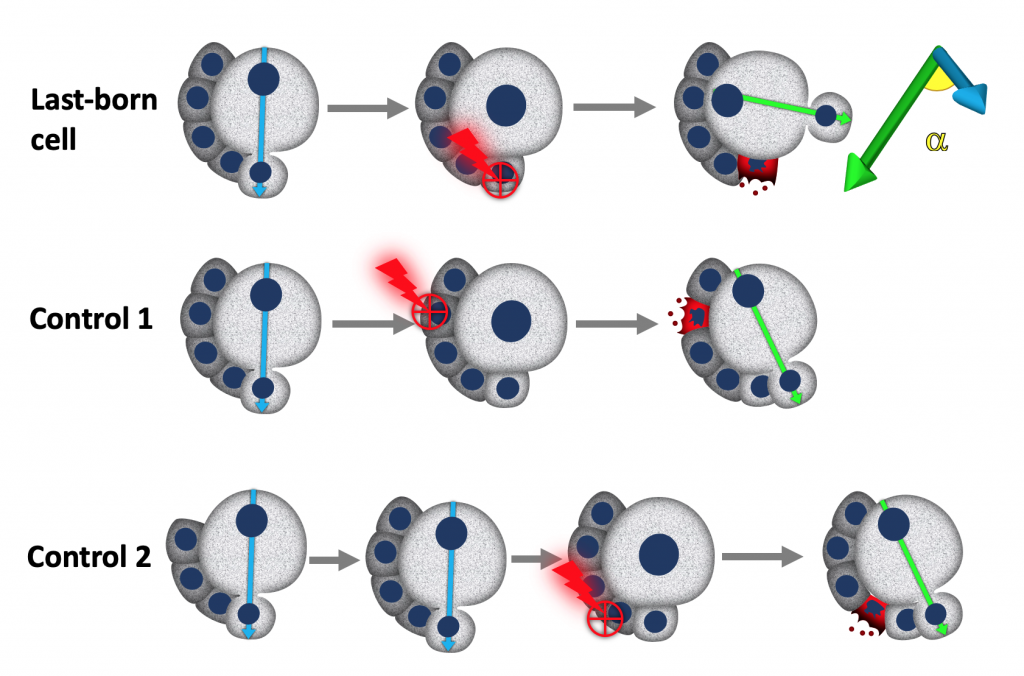
We used laser ablation in combination with live cell imaging and genetics to test the hypothesis that neuroblasts, may received cues from their own daughter cells that specify the orientation of the polarity axis and consequently the orientation of their division. Indeed, Neuroblast seem to be able to read a cue depending on their last-born daughter cells, which we propose o be important to maximise the contact of neuroblasts with neighbouring glial cells, that provide protection against environmental stresses.
How do cells position the proteins within them?
How cells position their proteins is a key problem in cell biology. Localizing mRNAs to distinct regions of the cytoplasm coupled to local activation of translation is one way to position proteins. mRNAs can also function as scaffolds to position proteins. We tested if mRNA localization contributes to the polarised distribution of proteins in mitotic neuroblasts to shed light on the molecular mechanism of cell fate generation in this system.
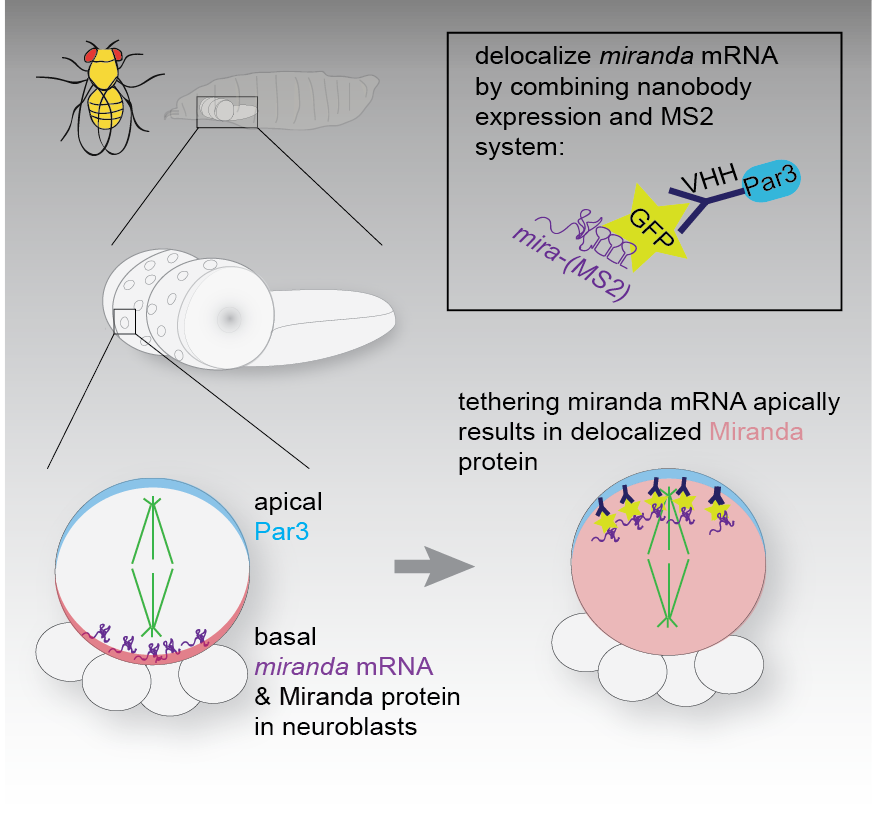
To this end we tagged endogenous mRNA using CrispR with GFP (via the MS2 system), to be able to visualise the localisation of mRNAs of candidate genes in living neuroblasts. We wanted to understand if the localisation of mRNA of genes involved in asymmetric neuroblast division, provided spatial information for the localisation of the encoded protein. To test this we altered the localisation of the GFP-tagged mRNA by expressing GFP-specific nanobodies fused to specific subcellular localization domains, which sequestered the mRNA in ectopic locations. In Ramat et al. we report that the mRNA of Miranda like the protein it encodes, is asymmetrically localized in mitotic neuroblast. Altering the sub cellular localization of the mRNA did not affect its translation, but resulted in mislocalization of the encoded protein. Protein localization defects caused by mislocalization of the cognate mRNA were rescued by introducing untagged mRNA coding for mutant non-localizable protein. Thus, the MS2 system and nanobody expression can be combined to redirect subcellular mRNA localization. In the case of miranda, reveal that interaction of Miranda protein and its mRNA is required for maintenance of the subcellular localization of Mira protein.
Quantitative live cell imaging
Neuroblasts in the Drosophila brain can be readily and without ambiguity identified, are emanable to long-term live cell imaging and can be manipulated using genetics or pharmacological approaches. In addition they are highly proliferative and can divide once every hour. These features make this system ideal to extract quantitive information in vivo and in real time to reveal the dynamic nature of the molecular processes that drive asymmetric cell division and cell fate specification in this system. We use this system to study the establishment and maintenance of cell polarity, a prerequisite of cell fate determination in this system. The evolutionarily conserved PAR polarity complex is required to establish polarity in neuroblasts. Therefore this system is ideal to reveal basic principles of how this important polarity complex functions and is regulated.
Chemical genetics in Drosophila
One caveat of the highly proliferative nature of neuroblasts is that standard genetic approaches such as RNAi or the use of mutant alleles can not easily if at all provide when in time a particular protein or enzyme is required. We are interested in how atypical protein kinase C (aPKC) operates in neuroblast to pattern the localization of cell fate determining molecules in mitosis. We recently developed the used of chemical genetics, an approach originally developed by the Shokat lab at UCSF in Drosophila.
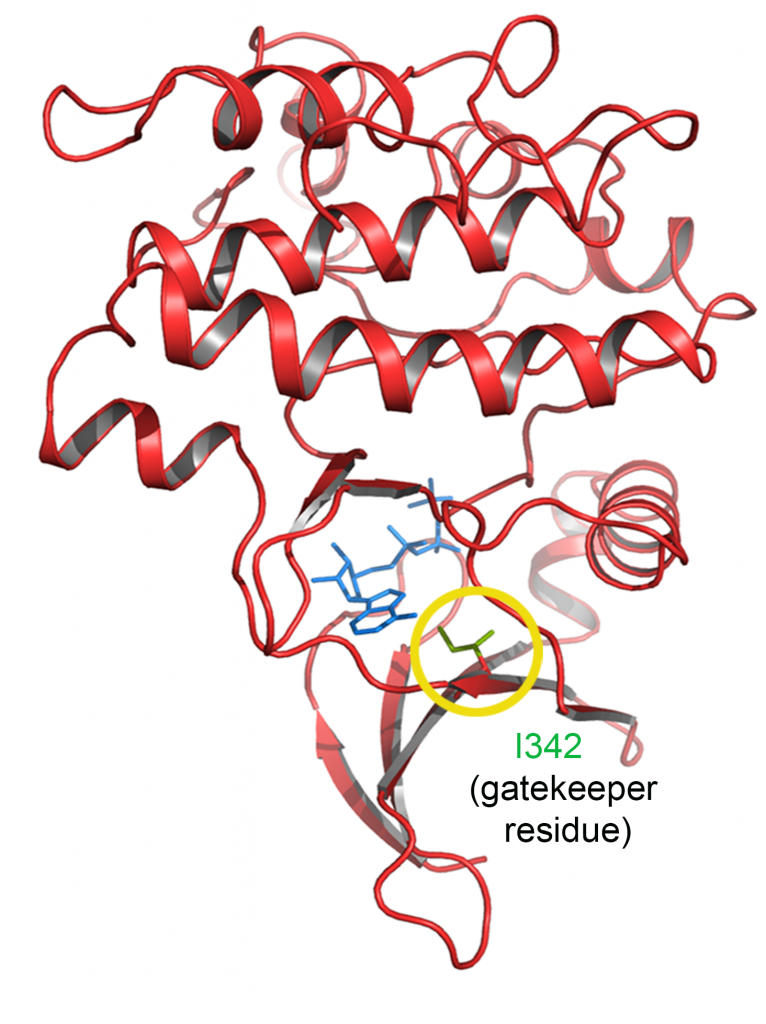
We used CrispR/Cas to engineer an analog sensitive allele of aPKC that can be specifically inhibited within minutes. Using this tool, we were able to study the requirement of aPKC’s kinase activity at different stages in the neuroblast cell cycle to reveal elements of the molecular mechanism that pattern the localization of fate determinants. We are currently developing more analog-sensitive alleles of other kinases of interest.
The video shows Drosophila egg chambers expressing PAR3 (Bazooka) fused to GFP that were also stained with SIR-Actin to reveal the actin cytoskeleton. Par3 localization is known to depend on aPKC phosphorylation in this system. These egg chambers are homozygous for the analog sensitive allele of apkc and aPKC was acutely inhibited after the first few frames, leading to contraction of the actin cytoskeleton and Par3 localization defects, demonstrating that the chemical genetics approach works in vivo in Drosophila. (Merge/Par3/SIR-Actin)